multiplexed DIA: plexDIA
Increasing the throughput of sensitive proteomics by plexDIA
plexDIA Article plexDIA code on GitHub Single-cell plexDIA
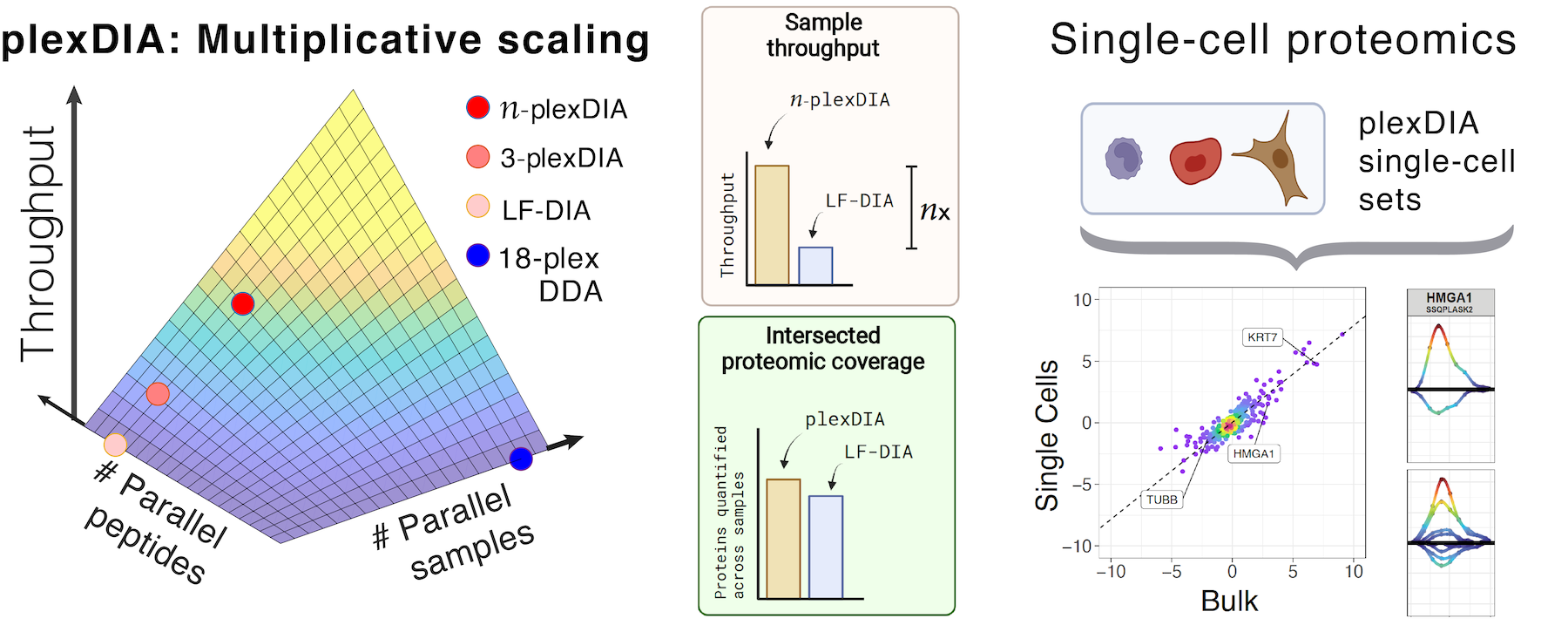
Current mass-spectrometry methods enable high-throughput proteomics of large sample amounts, but proteomics of low sample amounts remains limited in depth and throughput. To increase the throughput of sensitive proteomics, we developed an experimental and computational framework, plexDIA, for simultaneously multiplexing the analysis of both peptides and samples. Multiplexed analysis with plexDIA increases throughput multiplicatively with the number of labels without reducing proteome coverage or quantitative accuracy. By using 3-plex nonisobaric mass tags, plexDIA enables quantifying 3-fold more protein ratios among nanogram-level samples. Using 1 hour active gradients and first-generation Q Exactive, plexDIA quantified about 8,000 proteins in each sample of labeled 3-plex sets. plexDIA also increases data completeness, reducing missing data over 2-fold across samples. When applied to single human cells, plexDIA quantified about 1,000 proteins per cell and achieved 98 % data completeness within a plexDIA set while using about 5 min of active chromatography per cell. These results establish a general framework for increasing the throughput of sensitive and quantitative protein analysis.
- Derks, J., Leduc, A., et al. Increasing the throughput of sensitive proteomics by plexDIA. Nat Biotechnol (2022). 10.1038/s41587-022-01389-w, Preprint
Perspectives on high-throughput multiplexed proteomics
- Strategies for increasing the depth and throughput of protein analysis by plexDIA Journal of Proteome Research
- Framework for multiplicative scaling of single-cell proteomics, Nature Biotechnology
- Increasing proteomics throughput, Nature Biotechnology
- Driving Single Cell Proteomics Forward with Innovation, Journal of Proteome Research
- Scaling up single-cell proteomics, Molecular and Cellular Proteomics
About the project
plexDIA is a project developed in the Slavov Laboratory at Northeastern University in collaboration with Demichev and Rasler Laboratories at Charité, Universitätsmedizin. It was authored by Jason Derks, Andrew Leduc, Harrison Specht, R. Gray Huffman, Markus Ralser, Vadim Demichev and Nikolai Slavov.
Contact the authors by email: nslavov{at}northeastern.edu.
This project was supported by funding from the NIH Director’s Award and by an Allen Distinguished Investigator Award from the Paul G. Allen Frontiers Group.